Simulating a Point Source: Difference between revisions
(→Setting up the .xml's: added raikoke .xml) |
(Some minor changes) |
||
| (6 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
In this |
In this tutorial, the steps to simulate a volcanic eruption via a point source are given. |
||
[[File:Pointsource.gif]] |
|||
== Setting up ICON-ART == |
== Setting up ICON-ART == |
||
The first thing to do is having a working installation of ICON- |
The first thing to do is having a working installation of ICON-ART. To check if your icon version has been built correctly, you can check if |
||
your_icon_folder/bin/icon.exe |
your_icon_folder/bin/icon.exe |
||
exists. |
exists. |
||
| Line 12: | Line 13: | ||
* '''Working Directory''': Here the files that are needed for an individual run are saved. This usually includes the runscript and the relevant .xml files. |
* '''Working Directory''': Here the files that are needed for an individual run are saved. This usually includes the runscript and the relevant .xml files. |
||
* '''Icon Code directory''': This is where the Icon code is stored. |
* '''Icon Code directory''': This is where the Icon code is stored. |
||
* '''External Data directory''': Here external files which are needed for a run are stored. For example, to parametrize the optical properties of clouds a files like ECHAM6_CldOptProps.nc is used. These files of course can be switched out for others, however in most applications the same ones are used. A list of the used files is given in the |
* '''External Data directory''': Here external files which are needed for a run are stored. For example, to parametrize the optical properties of clouds a files like ECHAM6_CldOptProps.nc is used. These files of course can be switched out for others, however in most applications the same ones are used. A list of the used files is given in the runscript, which then creates a link of these files in the output directory. |
||
* '''Output directory''': This is where the new |
* '''Output directory''': This is where the new simulation data will be stored. Since most of the time large amounts of data are produced, this is stored in the work or scratch partitions on most HPC systems. The namelists produced by the runfile are also stored here. |
||
== Setting up the Runscript == |
== Setting up the Runscript == |
||
The function of the runscript is to set up the directory structure,link the relevant files and create the namelists. |
The function of the runscript is to set up the directory structure, link the relevant files and create the namelists, which are grouped into several namelist files. |
||
== Setting up the .xml's == |
== Setting up the .xml's == |
||
Here the .xml |
Here the .xml |
||
pntSRC.xml |
pntSRC.xml |
||
is set up. It contains all the necessary information to describe the emission of the here defined aerosols into the atmosphere. |
is set up. It contains all the necessary information to describe the emission of the here defined aerosols into the atmosphere. |
||
| Line 28: | Line 29: | ||
* what substances are emitted |
* what substances are emitted |
||
* how much of each substance is emitted |
* how much of each substance is emitted |
||
* what size are the emitted substances (median and standard deviation) |
* what size are the emitted substances (median and standard deviation) |
||
This can be adapted as needed. |
This can be adapted as needed. |
||
In this example |
In this example the Raikoke eruption on 21 of June 2019 is simulated, as can be seen in the following .xml: |
||
< |
<syntaxhighlight line lang=xml class="mw-collapsible-content"> |
||
<?xml version="1.0" encoding="UTF-8"?> |
<?xml version="1.0" encoding="UTF-8"?> |
||
| Line 69: | Line 70: | ||
<substance type="char">ash_insol_coa</substance> |
<substance type="char">ash_insol_coa</substance> |
||
<dg3_emiss type="real">2.98E-6</dg3_emiss> |
<dg3_emiss type="real">2.98E-6</dg3_emiss> |
||
<sigma_emiss type="real">1.4</sigma_emiss> |
<sigma_emiss type="real">1.4</sigma_emiss> |
||
<source_strength type="real">19700.0</source_strength> |
<source_strength type="real">19700.0</source_strength> |
||
<height type="real">14000</height> |
<height type="real">14000</height> |
||
| Line 91: | Line 92: | ||
</pntSrc> |
</pntSrc> |
||
</sources> |
</sources> |
||
</syntaxhighlight> |
|||
</pre> |
|||
== Running the Simulation == |
== Running the Simulation == |
||
The setup of the model for the simulation happens with a runscript. It contains all the different control parameters, such as time control of the simulation, what equations are going to be used, whether ART is active, and a lot more. |
|||
The runscript itself is just a text file, which is run via bash to start the simulation. Usually a template file is used and adapted to the specific needs of the user running the simulation. |
|||
Here we will go through the runscript for our pointsource simulation. |
|||
The first step is to set the relevant directories, so that the XML data we provide as well as the path to the ICON model can be found and accessed easily. |
|||
In this simulation, external data such as a grid containing external parameter data is required. The directory containing the data is set as DATADIR. |
|||
The output directory is where the model will write the output NetCDF files into, as well as linking a lot of relevant files used in the simulation. |
|||
<syntaxhighlight line lang=bash class="mw-collapsible-content"> |
|||
# working directory |
|||
export XMLDIR=/your/path/to/Training2023/Volcano |
|||
# datadir2 --> do not change |
|||
export DATADIR=/your/path/to/DATA_Volcano |
|||
# output directory |
|||
export OUTDIR=/your/path/to/Volcano_externally |
|||
# Code directory |
|||
export ICONDIR=/your/path/to/icon-kit |
|||
export ARTDIR=${ICONDIR}/externals/art |
|||
</syntaxhighlight> |
|||
In the next step, the previously defined output directory is created, if not already there, and the icon executable is linked into it for easier access and overview. |
|||
<syntaxhighlight line lang=bash class="mw-collapsible-content"> |
|||
if [ ! -d $OUTDIR ]; then |
|||
mkdir -p $OUTDIR |
|||
fi |
|||
cd $OUTDIR |
|||
# copy icon binary to OUTDIR |
|||
cp ${ICONDIR}/bin/icon icon.exe |
|||
</syntaxhighlight> |
|||
Now, the external parameters and chemistry files are linked into the output directory, so that ICON and the user can find it in the output directory |
|||
<syntaxhighlight line lang=bash class="mw-collapsible-content"> |
|||
ln -sf ${DATADIR}/icon_grid_0024_R02B06_G.nc ${OUTDIR}/iconR2B06-DOM01.nc |
|||
ln -sf ${DATADIR}/icon_grid_0024_R02B06_G-grfinfo.nc ${OUTDIR}/iconR2B06-DOM01-grfinfo.nc |
|||
ln -sf ${DATADIR}/icon_extpar_0024_R02B06_G_20200917_tiles.nc ${OUTDIR}/extpar_iconR2B06-DOM01.nc |
|||
ln -sf ${DATADIR}/icon_init_0024_R02B06_2019062112.nc ${OUTDIR}/igfff00000000 |
|||
ln -sf ${ICONDIR}/data/rrtmg_lw.nc ${OUTDIR}/rrtmg_lw.nc |
|||
ln -sf ${ICONDIR}/data/ECHAM6_CldOptProps.nc ${OUTDIR}/ECHAM6_CldOptProps.nc |
|||
# Dictionary for the mapping: DWD GRIB2 names <-> ICON internal names |
|||
ln -sf ${ICONDIR}/run/ana_varnames_map_file.txt ${OUTDIR}/map_file.ana |
|||
## chemistry |
|||
ln -sf ${ARTDIR}/runctrl_examples/init_ctrl/Simnoy2002.dat ${OUTDIR}/Simnoy2002.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/init_ctrl/Linoz2004Br.dat ${OUTDIR}/Linoz2004Br.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-aer.dat ${OUTDIR}/FJX_scat-aer.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_j2j.dat ${OUTDIR}/FJX_j2j.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_j2j_extended.dat ${OUTDIR}/FJX_j2j_extended.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-cld.dat ${OUTDIR}/FJX_scat-cld.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-ssa.dat ${OUTDIR}/FJX_scat-ssa.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-UMa.dat ${OUTDIR}/FJX_scat-UMa.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec_extended.dat ${OUTDIR}/FJX_spec_extended.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec_extended_lyman.dat ${OUTDIR}/FJX_spec_extended_lyman.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec.dat ${OUTDIR}/FJX_spec.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/atmos_std.dat ${OUTDIR}/atmos_std.dat |
|||
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/atmos_h2och4.dat ${OUTDIR}/atmos_h2och4.dat |
|||
</syntaxhighlight> |
|||
In the next step, namelist files are created. They contain the individual namelists which control the settings of the different modules of icon. They are written into the output directory and read by icon during a run. |
|||
First the master namelist is created, which most importantly contains the start and stop date, as well as the possibility to restart a run. |
|||
The next namelist file created is called "NAMELIST_Raikoke-June-2019". Here a lot of the model setup is taking place. It is subdivided into different namelists, which all do their part to set up a different part of the model. |
|||
Some important namelists for this simulation are: |
|||
* grid_nml (line 35): Here the grid on which the simulation is taking place is given |
|||
* run_nml (line 49): Contains the switch to turn on ICON-ART, as well as set model height levels and timestep length. |
|||
* output_nml (line 225): Defines which variables at which intervals are written in the output directory |
|||
* art_nml (line 238): Contains the switches relevant for ICON-ART, see [[Namelist]] for more details. Most importantly, the xml file for the point source emission is linked here |
|||
<syntaxhighlight line lang=bash class="mw-collapsible-content"> |
|||
cd $OUTDIR |
|||
cat > icon_master.namelist << EOF |
|||
&master_nml |
|||
lRestart = .false. |
|||
/ |
|||
&master_time_control_nml |
|||
experimentStartDate = "2019-06-21T12:00:00" |
|||
forecastLeadTime = "P1D" |
|||
checkpointTimeIntval = "P10D" |
|||
/ |
|||
&master_model_nml |
|||
model_type=1 |
|||
model_name="ATMO" |
|||
model_namelist_filename="NAMELIST_Raikoke-June-2019" |
|||
model_min_rank=1 |
|||
model_max_rank=65536 |
|||
model_inc_rank=1 |
|||
/ |
|||
&time_nml |
|||
ini_datetime_string = "2019-06-21T12:00:00" |
|||
dt_restart = 864000 ! 10 days |
|||
/ |
|||
EOF |
|||
cat > NAMELIST_Raikoke-June-2019 << EOF |
|||
¶llel_nml |
|||
nproma = 8 ! optimal setting 8 for CRAY; use 16 or 24 for IBM |
|||
p_test_run = .false. |
|||
l_test_openmp = .false. |
|||
l_log_checks = .false. |
|||
num_io_procs = 1 ! up to one PE per output stream is possible |
|||
itype_comm = 1 |
|||
iorder_sendrecv = 3 ! best value for CRAY (slightly faster than option 1) |
|||
/ |
|||
&grid_nml |
|||
dynamics_grid_filename = 'iconR2B06-DOM01.nc' |
|||
dynamics_parent_grid_id = 0 |
|||
lredgrid_phys = .false. |
|||
/ |
|||
&initicon_nml |
|||
init_mode = 7 ! 2: IFS; 7: Initialized DWD Analysis |
|||
lread_ana = .FALSE. ! no analysis data will be read |
|||
dwdfg_filename = "igfff00000000" ! initial data filename |
|||
filetype = 4 |
|||
ana_varnames_map_file = "map_file.ana" ! dictionary mapping internal names onto GRIB2 shortNames |
|||
ltile_coldstart = .TRUE. ! coldstart for surface tiles |
|||
ltile_init = .FALSE. ! set it to .TRUE. if FG data originate from run without tiles |
|||
/ |
|||
&run_nml |
|||
num_lev = 90 |
|||
lvert_nest = .true. ! use vertical nesting if a nest is active |
|||
!nsteps = ${nsteps} ! 50 ! 1200 ! 7200 ! |
|||
dtime = 180 ! timestep in seconds |
|||
ldynamics = .TRUE. ! dynamics |
|||
ltransport = .true. |
|||
iforcing = 3 ! NWP forcing |
|||
ltestcase = .false. ! false: run with real data |
|||
msg_level = 7 ! print maximum wind speeds every 5 time steps |
|||
ltimer = .true. ! set .TRUE. for timer output |
|||
timers_level = 15 ! can be increased up to 10 for detailed timer output |
|||
output = "nml" |
|||
lart = .true. |
|||
/ |
|||
&nwp_phy_nml |
|||
inwp_gscp = 1 |
|||
inwp_convection = 1 |
|||
inwp_radiation = 4 |
|||
inwp_cldcover = 1 |
|||
inwp_turb = 1 |
|||
inwp_satad = 1 |
|||
inwp_sso = 1 |
|||
inwp_gwd = 1 |
|||
inwp_surface = 1 |
|||
icapdcycl = 3 ! apply CAPE modification to improve diurnalcycle over tropical land (optimizes NWP scores) |
|||
latm_above_top = .false., .true. ! the second entry refers to the nested domain (if present) |
|||
efdt_min_raylfric = 3600 ! 7200. |
|||
itype_z0 = 2 |
|||
icpl_aero_conv = 0 ! check meaning -> default 0 - off |
|||
icpl_aero_gscp = 0 ! check meaning -> default 0 - off |
|||
! resolution-dependent settings - please choose the appropriate one |
|||
dt_rad = 2160. |
|||
dt_conv = 720. |
|||
dt_sso = 1440. |
|||
dt_gwd = 1440. |
|||
mu_rain = 0.5 |
|||
rain_n0_factor = 0.1 |
|||
lshallowconv_only = .true. |
|||
lgrayzone_deepconv = .false. |
|||
/ |
|||
&nwp_tuning_nml |
|||
itune_albedo = 1 ! somewhat reduced albedo (w.r.t. MODIS data) over Sahara in order to reduce cold bias |
|||
tune_gkdrag = 0.09 |
|||
tune_gkwake = 1.8 |
|||
tune_gfrcrit = 0.333 |
|||
tune_dust_abs = 1. |
|||
tune_zvz0i = 1.1 |
|||
tune_box_liq_asy = 4.0 |
|||
tune_gust_factor = 7.0 |
|||
tune_rcucov = 0.075 |
|||
tune_rhebc_land = 0.825 |
|||
tune_zvz0i = 0.85 |
|||
icpl_turb_clc = 2 |
|||
max_calibfac_clcl = 2.0 |
|||
tune_box_liq = 0.04 |
|||
/ |
|||
&turbdiff_nml |
|||
tkhmin = 0.75 ! new default since rev. 16527 |
|||
tkmmin = 0.75 ! " |
|||
pat_len = 750. ! 750 in exp.nh_oper |
|||
c_diff = 0.2 |
|||
rat_sea = 0.8 ! ** new value since for v2.0.15; previously 8.0 ** ! ** new since r20191: 8.5 for R3B7, 8.0 for R2B6 in order to get similar temperature biases in the tropics ** |
|||
ltkesso = .true. |
|||
frcsmot = 0.2 ! these 2 switches together apply vertical smoothing of the TKE source terms |
|||
imode_frcsmot = 2 ! in the tropics (only), which reduces the moist bias in the tropical lower troposphere: |
|||
itype_sher = 3 |
|||
ltkeshs = .true. |
|||
a_hshr = 2.0 |
|||
alpha1 = 0.125 |
|||
icldm_turb = 1 ! found in exp.nh_oper ** new recommendation for v2.0.15 in conjunction with evaporation fix for grid-scale rain ** |
|||
rlam_heat = 10.0 |
|||
/ |
|||
&lnd_nml |
|||
ntiles = 3 !!! 1 for assimilation cycle and forecast |
|||
nlev_snow = 3 !!! 1 for assimilation cycle and forecast |
|||
lmulti_snow = .false. !!! .false. for assimilation cycle and forecast |
|||
itype_heatcond = 3 |
|||
idiag_snowfrac = 20 |
|||
lsnowtile = .true. !! later on .true. if GRIB encoding issues are solved |
|||
lseaice = .true. |
|||
llake = .true. |
|||
lprog_albsi = .true. |
|||
itype_lndtbl = 4 ! minimizes moist/cold bias in lower tropical troposphere |
|||
itype_evsl = 4 |
|||
itype_root = 2 |
|||
itype_trvg = 3 |
|||
cwimax_ml = 5.e-4 |
|||
c_soil = 1.25 |
|||
c_soil_urb = 0.5 |
|||
sstice_mode = 2 |
|||
/ |
|||
&radiation_nml |
|||
irad_o3 = 7 |
|||
irad_aero = 6 |
|||
albedo_type = 2 ! Modis albedo |
|||
vmr_co2 = 390.e-06 ! values representative for 2012 |
|||
vmr_ch4 = 1800.e-09 |
|||
vmr_n2o = 322.0e-09 |
|||
vmr_o2 = 0.20946 |
|||
vmr_cfc11 = 240.e-12 |
|||
vmr_cfc12 = 532.e-12 |
|||
direct_albedo_water = 3 |
|||
albedo_whitecap = 1 |
|||
ecrad_data_path = "${ICONDIR}/externals/ecrad/data" |
|||
/ |
|||
&nonhydrostatic_nml |
|||
iadv_rhotheta = 2 |
|||
ivctype = 2 |
|||
itime_scheme = 4 |
|||
exner_expol = 0.333 |
|||
vwind_offctr = 0.2 |
|||
damp_height = 44000. |
|||
rayleigh_coeff = 1 |
|||
lhdiff_rcf = .true. |
|||
divdamp_order = 24 ! for data assimilation runs, '2' provides extra-strong filtering of gravity waves |
|||
divdamp_type = 32 !!! optional: 2 for assimilation cycle if very strong gravity-wave filtering is desired |
|||
divdamp_fac = 0.004 |
|||
l_open_ubc = .false. |
|||
igradp_method = 3 |
|||
l_zdiffu_t = .true. |
|||
thslp_zdiffu = 0.02 |
|||
thhgtd_zdiffu = 125. |
|||
htop_moist_proc= 22500. |
|||
hbot_qvsubstep = 22500. ! use 19000. with R3B7, must be at least as large as htop_moist_proc |
|||
htop_aero_proc = 25000. !height limit for aerosol tracer processes |
|||
/ |
|||
&sleve_nml |
|||
min_lay_thckn = 20. |
|||
max_lay_thckn = 400. ! maximum layer thickness below htop_thcknlimit |
|||
htop_thcknlimit = 14000. ! this implies that the upcoming COSMO-EU nest will have 60 levels |
|||
top_height = 75000. |
|||
stretch_fac = 0.9 |
|||
decay_scale_1 = 4000. |
|||
decay_scale_2 = 2500. |
|||
decay_exp = 1.2 |
|||
flat_height = 16000. |
|||
/ |
|||
&dynamics_nml |
|||
iequations = 3 |
|||
idiv_method = 1 |
|||
divavg_cntrwgt = 0.50 |
|||
lcoriolis = .TRUE. |
|||
/ |
|||
&transport_nml |
|||
ivadv_tracer = 3,3,3,3,3,3,3,3,3,3,3 |
|||
itype_hlimit = 3,4,4,4,4,3,3,3,3,3,3 |
|||
ihadv_tracer = 52,2,2,2,2,22,22,22,22,22,22 |
|||
iadv_tke = 0 |
|||
/ |
|||
&diffusion_nml |
|||
hdiff_order = 5 |
|||
itype_vn_diffu = 1 |
|||
itype_t_diffu = 2 |
|||
hdiff_efdt_ratio = 24.0 |
|||
hdiff_smag_fac = 0.025 |
|||
lhdiff_vn = .TRUE. |
|||
lhdiff_temp = .TRUE. |
|||
/ |
|||
&interpol_nml |
|||
nudge_zone_width = 8 |
|||
lsq_high_ord = 3 |
|||
l_intp_c2l = .true. |
|||
l_mono_c2l = .true. |
|||
/ |
|||
&extpar_nml |
|||
itopo = 1 |
|||
n_iter_smooth_topo = 1 |
|||
heightdiff_threshold = 3000. |
|||
hgtdiff_max_smooth_topo = 750., ! found in exp.nh_oper ** should be changed to 750.,750 with next Extpar update! ** |
|||
itype_lwemiss = 2 |
|||
/ |
|||
&io_nml |
|||
itype_pres_msl = 5 ! 5: new DWD method; 4: IFS method with bug fix for self-consistency between SLP and geopotential |
|||
itype_rh = 1 ! RH w.r.t. water |
|||
/ |
|||
&output_nml |
|||
filetype = 4 ! output format: 2=GRIB2, 4=NETCDFv2 |
|||
dom = -1 ! write all domains |
|||
mode = 1 ! 1 = forecast mode with TAXIS_RELATIVE, works only with output in multiples of 1h; 2 = climate mode, default, TAXIS_ABSOLUTE |
|||
output_bounds = 0., 691200., 600. ! start, end, increment |
|||
steps_per_file = 1 |
|||
include_last = .TRUE. |
|||
output_filename = 'Raikoke-June-2019-forecast_mode-remap' ! file name base |
|||
ml_varlist = 'TRSO2_chemtr','TRH2SO4_chemtr','group:ART_AEROSOL','rho','pres','temp', 'z_mc','z_ifc' |
|||
remap = 1 |
|||
reg_lon_def = 120.,0.1,280. |
|||
reg_lat_def = 85.,-0.1, 30. |
|||
/ |
|||
&art_nml |
|||
lart_diag_out = .TRUE. |
|||
lart_aerosol = .TRUE. |
|||
lart_pntSrc = .TRUE. |
|||
lart_chem = .TRUE. |
|||
lart_chemtracer = .TRUE. |
|||
iart_ari = 0 |
|||
cart_aerosol_xml = '${XMLDIR}/Ex01_aerotracer.xml' |
|||
cart_modes_xml = '${XMLDIR}/Ex01_modes.xml' |
|||
cart_pntSrc_xml = '${XMLDIR}/Ex01_pntSrc.xml' |
|||
cart_chemtracer_xml = '${XMLDIR}/Ex01_chemtracer.xml' |
|||
/ |
|||
EOF |
|||
</syntaxhighlight> |
|||
With the namelists set, we can now prepare the simulation for running. Since a lot of setup is required for running, this is also prepared within the runscript and then later executed. The following part is highly dependant on the HPC system configuration. The following setup works on the DKRZ Levante HPC system (July 2023): |
|||
<syntaxhighlight line lang=bash class="mw-collapsible-content"> |
|||
cat > ${OUTDIR}/job_ICON << ENDFILE |
|||
#!/bin/bash |
|||
#SBATCH --job-name=EXP01_ART # Specify job name |
|||
#SBATCH --partition=compute # Specify partition name |
|||
#SBATCH --nodes=4 # Specify number of nodes |
|||
#SBATCH --ntasks-per-node=128 # Specify number of (MPI) tasks on each node |
|||
#SBATCH --time=01:00:00 # Set a limit on the total run time |
|||
#SBATCH --mail-type=FAIL # Notify user by email in case of job failure |
|||
#SBATCH --mail-user=example@mail.com # Set your e-mail address |
|||
#SBATCH --account=your_project_account # Charge resources on this project account |
|||
module load gcc |
|||
module load intel-oneapi-compilers/2022.0.1-gcc-11.2.0 |
|||
module load intel-oneapi-mkl/2022.0.1-gcc-11.2.0 |
|||
module load openmpi/4.1.2-intel-2021.5.0 |
|||
module load netcdf-c/4.8.1-openmpi-4.1.2-intel-2021.5.0 |
|||
module load netcdf-fortran/4.5.3-openmpi-4.1.2-intel-2021.5.0 |
|||
module load parallel-netcdf/1.12.2-openmpi-4.1.2-intel-2021.5.0 |
|||
module load hdf5/1.12.1-openmpi-4.1.2-intel-2021.5.0 |
|||
module load eccodes/2.21.0-intel-2021.5.0 |
|||
unset SLURM_EXPORT_ENV |
|||
unset SLURM_MEM_PER_NODE |
|||
unset SBATCH_EXPORT |
|||
export LD_LIBRARY_PATH="/usr/lib:/usr/lib64:/sw/spack-levante/netcdf-c-4.8.1-2k3cmu/lib:/sw/spack-levante/netcdf-fortran-4.5.3-k6xq5g/lib:/sw/spack-levante/hdf5-1.12.1-tvymb5/lib:/sw/spack-levante/eccodes-2.21.0-3ehkbb/lib64:/sw/spack-levante/intel-oneapi-mkl-2022.0.1-ttdktf/mkl/2022.0.1/lib/intel64/" |
|||
export OMPI_MCA_pml="ucx" |
|||
export OMPI_MCA_btl=self |
|||
export OMPI_MCA_osc="pt2pt" |
|||
export UCX_IB_ADDR_TYPE=ib_global |
|||
export OMPI_MCA_coll="^ml,hcoll" |
|||
export OMPI_MCA_coll_hcoll_enable="0" |
|||
export HCOLL_ENABLE_MCAST_ALL="0" |
|||
export HCOLL_MAIN_IB=mlx5_0:1 |
|||
export UCX_NET_DEVICES=mlx5_0:1 |
|||
export UCX_TLS=mm,knem,cma,dc_mlx5,dc_x,self |
|||
export UCX_UNIFIED_MODE=y |
|||
export HDF5_USE_FILE_LOCKING=FALSE |
|||
export OMPI_MCA_io="romio321" |
|||
export UCX_HANDLE_ERRORS=bt |
|||
ulimit -s 102400 |
|||
ulimit -c 0 |
|||
srun -l --cpu_bind=cores --distribution=block:cyclic --propagate=STACK,CORE ./icon.exe |
|||
ENDFILE |
|||
</syntaxhighlight> |
|||
This concludes the runfile. |
|||
The Job_ICON-file that has been produced in the last step can then be executed. For that, the runfile hast to be run using bash: |
|||
<syntaxhighlight lang=bash class="mw-collapsible-content"> |
|||
$ chmod +x myrunfile |
|||
$ ./myrunfile |
|||
</syntaxhighlight> |
|||
This will then create all the Namelist files and the runscript job_ICON. To submit the icon run, go to the output directory, make the runscript executable, and run it: |
|||
<syntaxhighlight lang=bash class="mw-collapsible-content"> |
|||
$ cd /path/to/your/output |
|||
$ chmod +x job_ICON |
|||
$ sbatch job_ICON |
|||
</syntaxhighlight> |
|||
Now the Job should be submitted. The submission status can be seen with |
|||
<syntaxhighlight lang=bash class="mw-collapsible-content"> |
|||
$ squeue --user your_username |
|||
</syntaxhighlight> |
|||
If everything went well, in the output directory there will be output files generated with names like Raikoke-June-2019-forecast_mode-remap_DOM01_ML_0001.nc |
|||
== Inspecting the Output == |
== Inspecting the Output == |
||
In this example, the output is created in the form of several NetCDF4 (.nc) files. To avoid having one giant file, it is subdivided into several files containing a fixed amount of timesteps. The quickest way to inspect a NetCDF file is with [https://code.mpimet.mpg.de/projects/cdo CDO]: |
|||
<syntaxhighlight lang=bash class="mw-collapsible-content"> |
|||
$ cdo sinfo Raikoke-June-2019-forecast_mode-remap_DOM01_ML_0001.nc |
|||
</syntaxhighlight> |
|||
This way it is possible to obtain information about the variables, grid parameters and timesteps of the simulation. To actually see the values of a specific variable, tools like ncdump can be used, however in many cases this is not practical, since the amount of single values is way too large to look at individual data points. |
|||
== Making a Plot of the Simulation Data == |
== Making a Plot of the Simulation Data == |
||
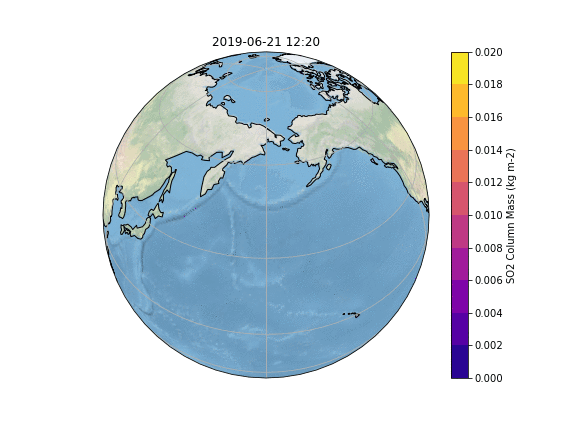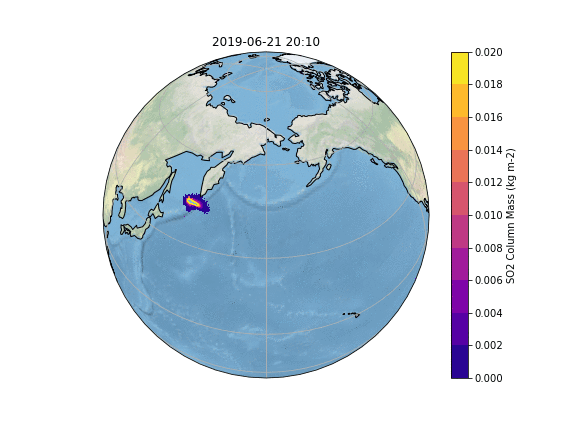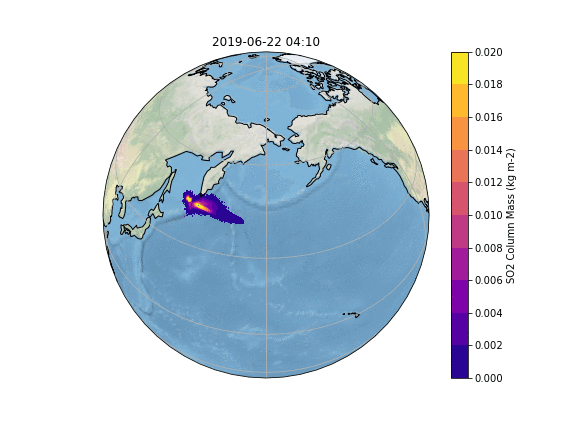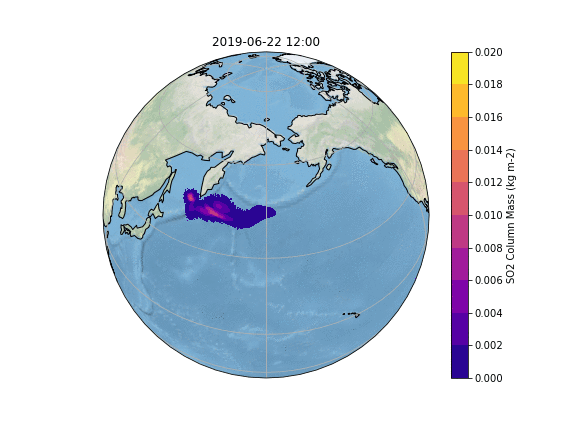
In order to gain insight into the simulation, as well as an overview, the most common way is to create some plots of different variables of interest. In our case, we want to know what happens to the ash emitted from the Raikoke eruption. |
|||
A quick and easy way to create plots is using python and a reference script to produce a plot similar to the Pointsource animation is given here: |
|||
<syntaxhighlight line lang=python class="mw-collapsible-content"> |
|||
# first we import the following packages: |
|||
import numpy as np # general utility for trigonomic functions and arrays |
|||
import matplotlib.pyplot as plt # Library to produce Plots |
|||
import xarray as xr # reading in of netCDF data, optimized for large datasets |
|||
from glob import glob |
|||
import cartopy.crs as ccrs # plotting things on a map |
|||
#Next step: read in the Netcdf Files |
|||
#datadir_externally = 'path/to/your/output/Training2023/Volcano_externally/' |
|||
files_externally = glob(datadir_externally+'Raikoke-June-2019-forecast_mode-remap_DOM01_ML*') |
|||
files_externally.sort() |
|||
#Next step: Calculate relevant data to be displayed |
|||
# get model level height |
|||
def get_dz(ds): |
|||
dz = -1 * ds.z_ifc.diff('height_2') |
|||
dz = dz.rename({'height_2':'height'}) |
|||
dz = dz.assign_coords(height=(dz.height - 1)) |
|||
return dz |
|||
# Calculate column integrated tracer mass (tracer load) |
|||
def tracer_load(tr,rho,dz): |
|||
m_tr = tr * rho * dz |
|||
m_tr = m_tr.sum('height') |
|||
m_tr = m_tr.squeeze() |
|||
return m_tr |
|||
# |
|||
MM_SO2 = 64/28.96 # molar weight of SO2 / dry air |
|||
# set the timestep you want to plot |
|||
# in this simulation there are 144 timesteps and 144 files each containing one timestep |
|||
# To produce a plot for each timestep, you can also loop the rest of this file over i |
|||
i = 30 |
|||
#load the NetCDF file containing the timestep |
|||
ds = xr.open_dataset(files_externally[i],autoclose=True) |
|||
#calculate Heightlevels |
|||
dz = get_dz(ds) |
|||
# Read in SO2 Column |
|||
SO2 = tracer_load(ds.TRSO2_chemtr.values*MM_SO2,ds.rho,dz) |
|||
# set colorbar levels |
|||
color_so2 = np.linspace(1e-5, 0.02, 11,endpoint=True) |
|||
# Plot SO2 column |
|||
# set the size of the figure and the background color to white |
|||
fig = plt.figure(figsize=(8,6),facecolor='w') |
|||
# Set the axes using the specified map projection |
|||
ax=plt.axes(projection=ccrs.NearsidePerspective(central_longitude=180, central_latitude=49.29,satellite_height=35785831/5, globe=None)) |
|||
ax.stock_img() #add a stock image as background |
|||
ax.coastlines() # add coastlines |
|||
ax.set_title(str(ds.time.values)[2:12]+" "+str(ds.time.values)[13:18]) #set title to the current time displayed |
|||
ax.gridlines(draw_labels=False, dms=False, x_inline=True, y_inline=True) #add lat lon gridlines |
|||
# Make a filled contour plot |
|||
plot1=ax.contourf(ds.lon, ds.lat, SO2, levels=color_so2, cmap='plasma', |
|||
transform = ccrs.PlateCarree()) |
|||
kwargs = {'format': '%.3f'} |
|||
cbar = plt.colorbar(plot1,location='right',ticks=color_so2,**kwargs) |
|||
cbar.set_label('SO2 Column Mass (kg m-2)') |
|||
#save the plot at a path of your choice |
|||
#plt.savefig("path/to/your/Plots/folder/SO2_{:03d}.png".format(i)) |
|||
plt.close(fig) |
|||
</syntaxhighlight> |
|||
To run the script, you need a working python version and the packages listed in the beginning of the script. Dont forget to make the file executable in order to run it using <code> chmod +x plotscript </code>. |
|||
The resulting Plot will look something like this: [[File:Example SO2 130.png]] |
|||
Latest revision as of 09:18, 26 October 2023
In this tutorial, the steps to simulate a volcanic eruption via a point source are given.
Setting up ICON-ART
The first thing to do is having a working installation of ICON-ART. To check if your icon version has been built correctly, you can check if
your_icon_folder/bin/icon.exe
exists.
Setting up Directories
Now a directory structure has to be set up. Usually the following directories are used:
- Working Directory: Here the files that are needed for an individual run are saved. This usually includes the runscript and the relevant .xml files.
- Icon Code directory: This is where the Icon code is stored.
- External Data directory: Here external files which are needed for a run are stored. For example, to parametrize the optical properties of clouds a files like ECHAM6_CldOptProps.nc is used. These files of course can be switched out for others, however in most applications the same ones are used. A list of the used files is given in the runscript, which then creates a link of these files in the output directory.
- Output directory: This is where the new simulation data will be stored. Since most of the time large amounts of data are produced, this is stored in the work or scratch partitions on most HPC systems. The namelists produced by the runfile are also stored here.
Setting up the Runscript
The function of the runscript is to set up the directory structure, link the relevant files and create the namelists, which are grouped into several namelist files.
Setting up the .xml's
Here the .xml
pntSRC.xml
is set up. It contains all the necessary information to describe the emission of the here defined aerosols into the atmosphere. The following information is contained:
- where is the Pointsource
- when is it emitting
- what substances are emitted
- how much of each substance is emitted
- what size are the emitted substances (median and standard deviation)
This can be adapted as needed.
In this example the Raikoke eruption on 21 of June 2019 is simulated, as can be seen in the following .xml:
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE tracers SYSTEM "sources_selTrnsp.dtd">
<sources>
<pntSrc id="Raikoke-SO2">
<lon type="real">153.24</lon>
<lat type="real">49.29</lat>
<substance type="char">TRSO2</substance>
<source_strength type="real">46300.0</source_strength>
<height type="real">14000</height>
<height_bot type="real">600</height_bot>
<unit type="char">kg s-1</unit>
<startTime type="char">2019-06-21T13:00:00</startTime>
<endTime type="char">2019-06-21T22:00:00</endTime>
</pntSrc>
<pntSrc id="Raikoke-ashacc">
<lon type="real">153.24</lon>
<lat type="real">49.29</lat>
<substance type="char">ash_insol_acc</substance>
<dg3_emiss type="real">0.8E-6</dg3_emiss>
<sigma_emiss type="real">1.4</sigma_emiss>
<source_strength type="real">19700.0</source_strength>
<height type="real">14000</height>
<height_bot type="real">600</height_bot>
<unit type="char">kg s-1</unit>
<startTime type="char">2019-06-21T13:00:00</startTime>
<endTime type="char">2019-06-21T22:00:00</endTime>
</pntSrc>
<pntSrc id="Raikoke-ashcoa">
<lon type="real">153.24</lon>
<lat type="real">49.29</lat>
<substance type="char">ash_insol_coa</substance>
<dg3_emiss type="real">2.98E-6</dg3_emiss>
<sigma_emiss type="real">1.4</sigma_emiss>
<source_strength type="real">19700.0</source_strength>
<height type="real">14000</height>
<height_bot type="real">600</height_bot>
<unit type="char">kg s-1</unit>
<startTime type="char">2019-06-21T13:00:00</startTime>
<endTime type="char">2019-06-21T22:00:00</endTime>
</pntSrc>
<pntSrc id="Raikoke-ashgiant">
<lon type="real">153.24</lon>
<lat type="real">49.29</lat>
<substance type="char">ash_giant</substance>
<dg3_emiss type="real">11.35E-6</dg3_emiss>
<sigma_emiss type="real">1.4</sigma_emiss>
<source_strength type="real">19700.0</source_strength>
<height type="real">14000</height>
<height_bot type="real">600</height_bot>
<unit type="char">kg s-1</unit>
<startTime type="char">2019-06-21T13:00:00</startTime>
<endTime type="char">2019-06-21T22:00:00</endTime>
</pntSrc>
</sources>
Running the Simulation
The setup of the model for the simulation happens with a runscript. It contains all the different control parameters, such as time control of the simulation, what equations are going to be used, whether ART is active, and a lot more. The runscript itself is just a text file, which is run via bash to start the simulation. Usually a template file is used and adapted to the specific needs of the user running the simulation. Here we will go through the runscript for our pointsource simulation.
The first step is to set the relevant directories, so that the XML data we provide as well as the path to the ICON model can be found and accessed easily. In this simulation, external data such as a grid containing external parameter data is required. The directory containing the data is set as DATADIR. The output directory is where the model will write the output NetCDF files into, as well as linking a lot of relevant files used in the simulation.
# working directory
export XMLDIR=/your/path/to/Training2023/Volcano
# datadir2 --> do not change
export DATADIR=/your/path/to/DATA_Volcano
# output directory
export OUTDIR=/your/path/to/Volcano_externally
# Code directory
export ICONDIR=/your/path/to/icon-kit
export ARTDIR=${ICONDIR}/externals/art
In the next step, the previously defined output directory is created, if not already there, and the icon executable is linked into it for easier access and overview.
if [ ! -d $OUTDIR ]; then
mkdir -p $OUTDIR
fi
cd $OUTDIR
# copy icon binary to OUTDIR
cp ${ICONDIR}/bin/icon icon.exe
Now, the external parameters and chemistry files are linked into the output directory, so that ICON and the user can find it in the output directory
ln -sf ${DATADIR}/icon_grid_0024_R02B06_G.nc ${OUTDIR}/iconR2B06-DOM01.nc
ln -sf ${DATADIR}/icon_grid_0024_R02B06_G-grfinfo.nc ${OUTDIR}/iconR2B06-DOM01-grfinfo.nc
ln -sf ${DATADIR}/icon_extpar_0024_R02B06_G_20200917_tiles.nc ${OUTDIR}/extpar_iconR2B06-DOM01.nc
ln -sf ${DATADIR}/icon_init_0024_R02B06_2019062112.nc ${OUTDIR}/igfff00000000
ln -sf ${ICONDIR}/data/rrtmg_lw.nc ${OUTDIR}/rrtmg_lw.nc
ln -sf ${ICONDIR}/data/ECHAM6_CldOptProps.nc ${OUTDIR}/ECHAM6_CldOptProps.nc
# Dictionary for the mapping: DWD GRIB2 names <-> ICON internal names
ln -sf ${ICONDIR}/run/ana_varnames_map_file.txt ${OUTDIR}/map_file.ana
## chemistry
ln -sf ${ARTDIR}/runctrl_examples/init_ctrl/Simnoy2002.dat ${OUTDIR}/Simnoy2002.dat
ln -sf ${ARTDIR}/runctrl_examples/init_ctrl/Linoz2004Br.dat ${OUTDIR}/Linoz2004Br.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-aer.dat ${OUTDIR}/FJX_scat-aer.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_j2j.dat ${OUTDIR}/FJX_j2j.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_j2j_extended.dat ${OUTDIR}/FJX_j2j_extended.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-cld.dat ${OUTDIR}/FJX_scat-cld.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-ssa.dat ${OUTDIR}/FJX_scat-ssa.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_scat-UMa.dat ${OUTDIR}/FJX_scat-UMa.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec_extended.dat ${OUTDIR}/FJX_spec_extended.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec_extended_lyman.dat ${OUTDIR}/FJX_spec_extended_lyman.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/FJX_spec.dat ${OUTDIR}/FJX_spec.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/atmos_std.dat ${OUTDIR}/atmos_std.dat
ln -sf ${ARTDIR}/runctrl_examples/photo_ctrl/atmos_h2och4.dat ${OUTDIR}/atmos_h2och4.dat
In the next step, namelist files are created. They contain the individual namelists which control the settings of the different modules of icon. They are written into the output directory and read by icon during a run. First the master namelist is created, which most importantly contains the start and stop date, as well as the possibility to restart a run. The next namelist file created is called "NAMELIST_Raikoke-June-2019". Here a lot of the model setup is taking place. It is subdivided into different namelists, which all do their part to set up a different part of the model. Some important namelists for this simulation are:
- grid_nml (line 35): Here the grid on which the simulation is taking place is given
- run_nml (line 49): Contains the switch to turn on ICON-ART, as well as set model height levels and timestep length.
- output_nml (line 225): Defines which variables at which intervals are written in the output directory
- art_nml (line 238): Contains the switches relevant for ICON-ART, see Namelist for more details. Most importantly, the xml file for the point source emission is linked here
cd $OUTDIR
cat > icon_master.namelist << EOF
&master_nml
lRestart = .false.
/
&master_time_control_nml
experimentStartDate = "2019-06-21T12:00:00"
forecastLeadTime = "P1D"
checkpointTimeIntval = "P10D"
/
&master_model_nml
model_type=1
model_name="ATMO"
model_namelist_filename="NAMELIST_Raikoke-June-2019"
model_min_rank=1
model_max_rank=65536
model_inc_rank=1
/
&time_nml
ini_datetime_string = "2019-06-21T12:00:00"
dt_restart = 864000 ! 10 days
/
EOF
cat > NAMELIST_Raikoke-June-2019 << EOF
¶llel_nml
nproma = 8 ! optimal setting 8 for CRAY; use 16 or 24 for IBM
p_test_run = .false.
l_test_openmp = .false.
l_log_checks = .false.
num_io_procs = 1 ! up to one PE per output stream is possible
itype_comm = 1
iorder_sendrecv = 3 ! best value for CRAY (slightly faster than option 1)
/
&grid_nml
dynamics_grid_filename = 'iconR2B06-DOM01.nc'
dynamics_parent_grid_id = 0
lredgrid_phys = .false.
/
&initicon_nml
init_mode = 7 ! 2: IFS; 7: Initialized DWD Analysis
lread_ana = .FALSE. ! no analysis data will be read
dwdfg_filename = "igfff00000000" ! initial data filename
filetype = 4
ana_varnames_map_file = "map_file.ana" ! dictionary mapping internal names onto GRIB2 shortNames
ltile_coldstart = .TRUE. ! coldstart for surface tiles
ltile_init = .FALSE. ! set it to .TRUE. if FG data originate from run without tiles
/
&run_nml
num_lev = 90
lvert_nest = .true. ! use vertical nesting if a nest is active
!nsteps = ${nsteps} ! 50 ! 1200 ! 7200 !
dtime = 180 ! timestep in seconds
ldynamics = .TRUE. ! dynamics
ltransport = .true.
iforcing = 3 ! NWP forcing
ltestcase = .false. ! false: run with real data
msg_level = 7 ! print maximum wind speeds every 5 time steps
ltimer = .true. ! set .TRUE. for timer output
timers_level = 15 ! can be increased up to 10 for detailed timer output
output = "nml"
lart = .true.
/
&nwp_phy_nml
inwp_gscp = 1
inwp_convection = 1
inwp_radiation = 4
inwp_cldcover = 1
inwp_turb = 1
inwp_satad = 1
inwp_sso = 1
inwp_gwd = 1
inwp_surface = 1
icapdcycl = 3 ! apply CAPE modification to improve diurnalcycle over tropical land (optimizes NWP scores)
latm_above_top = .false., .true. ! the second entry refers to the nested domain (if present)
efdt_min_raylfric = 3600 ! 7200.
itype_z0 = 2
icpl_aero_conv = 0 ! check meaning -> default 0 - off
icpl_aero_gscp = 0 ! check meaning -> default 0 - off
! resolution-dependent settings - please choose the appropriate one
dt_rad = 2160.
dt_conv = 720.
dt_sso = 1440.
dt_gwd = 1440.
mu_rain = 0.5
rain_n0_factor = 0.1
lshallowconv_only = .true.
lgrayzone_deepconv = .false.
/
&nwp_tuning_nml
itune_albedo = 1 ! somewhat reduced albedo (w.r.t. MODIS data) over Sahara in order to reduce cold bias
tune_gkdrag = 0.09
tune_gkwake = 1.8
tune_gfrcrit = 0.333
tune_dust_abs = 1.
tune_zvz0i = 1.1
tune_box_liq_asy = 4.0
tune_gust_factor = 7.0
tune_rcucov = 0.075
tune_rhebc_land = 0.825
tune_zvz0i = 0.85
icpl_turb_clc = 2
max_calibfac_clcl = 2.0
tune_box_liq = 0.04
/
&turbdiff_nml
tkhmin = 0.75 ! new default since rev. 16527
tkmmin = 0.75 ! "
pat_len = 750. ! 750 in exp.nh_oper
c_diff = 0.2
rat_sea = 0.8 ! ** new value since for v2.0.15; previously 8.0 ** ! ** new since r20191: 8.5 for R3B7, 8.0 for R2B6 in order to get similar temperature biases in the tropics **
ltkesso = .true.
frcsmot = 0.2 ! these 2 switches together apply vertical smoothing of the TKE source terms
imode_frcsmot = 2 ! in the tropics (only), which reduces the moist bias in the tropical lower troposphere:
itype_sher = 3
ltkeshs = .true.
a_hshr = 2.0
alpha1 = 0.125
icldm_turb = 1 ! found in exp.nh_oper ** new recommendation for v2.0.15 in conjunction with evaporation fix for grid-scale rain **
rlam_heat = 10.0
/
&lnd_nml
ntiles = 3 !!! 1 for assimilation cycle and forecast
nlev_snow = 3 !!! 1 for assimilation cycle and forecast
lmulti_snow = .false. !!! .false. for assimilation cycle and forecast
itype_heatcond = 3
idiag_snowfrac = 20
lsnowtile = .true. !! later on .true. if GRIB encoding issues are solved
lseaice = .true.
llake = .true.
lprog_albsi = .true.
itype_lndtbl = 4 ! minimizes moist/cold bias in lower tropical troposphere
itype_evsl = 4
itype_root = 2
itype_trvg = 3
cwimax_ml = 5.e-4
c_soil = 1.25
c_soil_urb = 0.5
sstice_mode = 2
/
&radiation_nml
irad_o3 = 7
irad_aero = 6
albedo_type = 2 ! Modis albedo
vmr_co2 = 390.e-06 ! values representative for 2012
vmr_ch4 = 1800.e-09
vmr_n2o = 322.0e-09
vmr_o2 = 0.20946
vmr_cfc11 = 240.e-12
vmr_cfc12 = 532.e-12
direct_albedo_water = 3
albedo_whitecap = 1
ecrad_data_path = "${ICONDIR}/externals/ecrad/data"
/
&nonhydrostatic_nml
iadv_rhotheta = 2
ivctype = 2
itime_scheme = 4
exner_expol = 0.333
vwind_offctr = 0.2
damp_height = 44000.
rayleigh_coeff = 1
lhdiff_rcf = .true.
divdamp_order = 24 ! for data assimilation runs, '2' provides extra-strong filtering of gravity waves
divdamp_type = 32 !!! optional: 2 for assimilation cycle if very strong gravity-wave filtering is desired
divdamp_fac = 0.004
l_open_ubc = .false.
igradp_method = 3
l_zdiffu_t = .true.
thslp_zdiffu = 0.02
thhgtd_zdiffu = 125.
htop_moist_proc= 22500.
hbot_qvsubstep = 22500. ! use 19000. with R3B7, must be at least as large as htop_moist_proc
htop_aero_proc = 25000. !height limit for aerosol tracer processes
/
&sleve_nml
min_lay_thckn = 20.
max_lay_thckn = 400. ! maximum layer thickness below htop_thcknlimit
htop_thcknlimit = 14000. ! this implies that the upcoming COSMO-EU nest will have 60 levels
top_height = 75000.
stretch_fac = 0.9
decay_scale_1 = 4000.
decay_scale_2 = 2500.
decay_exp = 1.2
flat_height = 16000.
/
&dynamics_nml
iequations = 3
idiv_method = 1
divavg_cntrwgt = 0.50
lcoriolis = .TRUE.
/
&transport_nml
ivadv_tracer = 3,3,3,3,3,3,3,3,3,3,3
itype_hlimit = 3,4,4,4,4,3,3,3,3,3,3
ihadv_tracer = 52,2,2,2,2,22,22,22,22,22,22
iadv_tke = 0
/
&diffusion_nml
hdiff_order = 5
itype_vn_diffu = 1
itype_t_diffu = 2
hdiff_efdt_ratio = 24.0
hdiff_smag_fac = 0.025
lhdiff_vn = .TRUE.
lhdiff_temp = .TRUE.
/
&interpol_nml
nudge_zone_width = 8
lsq_high_ord = 3
l_intp_c2l = .true.
l_mono_c2l = .true.
/
&extpar_nml
itopo = 1
n_iter_smooth_topo = 1
heightdiff_threshold = 3000.
hgtdiff_max_smooth_topo = 750., ! found in exp.nh_oper ** should be changed to 750.,750 with next Extpar update! **
itype_lwemiss = 2
/
&io_nml
itype_pres_msl = 5 ! 5: new DWD method; 4: IFS method with bug fix for self-consistency between SLP and geopotential
itype_rh = 1 ! RH w.r.t. water
/
&output_nml
filetype = 4 ! output format: 2=GRIB2, 4=NETCDFv2
dom = -1 ! write all domains
mode = 1 ! 1 = forecast mode with TAXIS_RELATIVE, works only with output in multiples of 1h; 2 = climate mode, default, TAXIS_ABSOLUTE
output_bounds = 0., 691200., 600. ! start, end, increment
steps_per_file = 1
include_last = .TRUE.
output_filename = 'Raikoke-June-2019-forecast_mode-remap' ! file name base
ml_varlist = 'TRSO2_chemtr','TRH2SO4_chemtr','group:ART_AEROSOL','rho','pres','temp', 'z_mc','z_ifc'
remap = 1
reg_lon_def = 120.,0.1,280.
reg_lat_def = 85.,-0.1, 30.
/
&art_nml
lart_diag_out = .TRUE.
lart_aerosol = .TRUE.
lart_pntSrc = .TRUE.
lart_chem = .TRUE.
lart_chemtracer = .TRUE.
iart_ari = 0
cart_aerosol_xml = '${XMLDIR}/Ex01_aerotracer.xml'
cart_modes_xml = '${XMLDIR}/Ex01_modes.xml'
cart_pntSrc_xml = '${XMLDIR}/Ex01_pntSrc.xml'
cart_chemtracer_xml = '${XMLDIR}/Ex01_chemtracer.xml'
/
EOF
With the namelists set, we can now prepare the simulation for running. Since a lot of setup is required for running, this is also prepared within the runscript and then later executed. The following part is highly dependant on the HPC system configuration. The following setup works on the DKRZ Levante HPC system (July 2023):
cat > ${OUTDIR}/job_ICON << ENDFILE
#!/bin/bash
#SBATCH --job-name=EXP01_ART # Specify job name
#SBATCH --partition=compute # Specify partition name
#SBATCH --nodes=4 # Specify number of nodes
#SBATCH --ntasks-per-node=128 # Specify number of (MPI) tasks on each node
#SBATCH --time=01:00:00 # Set a limit on the total run time
#SBATCH --mail-type=FAIL # Notify user by email in case of job failure
#SBATCH --mail-user=example@mail.com # Set your e-mail address
#SBATCH --account=your_project_account # Charge resources on this project account
module load gcc
module load intel-oneapi-compilers/2022.0.1-gcc-11.2.0
module load intel-oneapi-mkl/2022.0.1-gcc-11.2.0
module load openmpi/4.1.2-intel-2021.5.0
module load netcdf-c/4.8.1-openmpi-4.1.2-intel-2021.5.0
module load netcdf-fortran/4.5.3-openmpi-4.1.2-intel-2021.5.0
module load parallel-netcdf/1.12.2-openmpi-4.1.2-intel-2021.5.0
module load hdf5/1.12.1-openmpi-4.1.2-intel-2021.5.0
module load eccodes/2.21.0-intel-2021.5.0
unset SLURM_EXPORT_ENV
unset SLURM_MEM_PER_NODE
unset SBATCH_EXPORT
export LD_LIBRARY_PATH="/usr/lib:/usr/lib64:/sw/spack-levante/netcdf-c-4.8.1-2k3cmu/lib:/sw/spack-levante/netcdf-fortran-4.5.3-k6xq5g/lib:/sw/spack-levante/hdf5-1.12.1-tvymb5/lib:/sw/spack-levante/eccodes-2.21.0-3ehkbb/lib64:/sw/spack-levante/intel-oneapi-mkl-2022.0.1-ttdktf/mkl/2022.0.1/lib/intel64/"
export OMPI_MCA_pml="ucx"
export OMPI_MCA_btl=self
export OMPI_MCA_osc="pt2pt"
export UCX_IB_ADDR_TYPE=ib_global
export OMPI_MCA_coll="^ml,hcoll"
export OMPI_MCA_coll_hcoll_enable="0"
export HCOLL_ENABLE_MCAST_ALL="0"
export HCOLL_MAIN_IB=mlx5_0:1
export UCX_NET_DEVICES=mlx5_0:1
export UCX_TLS=mm,knem,cma,dc_mlx5,dc_x,self
export UCX_UNIFIED_MODE=y
export HDF5_USE_FILE_LOCKING=FALSE
export OMPI_MCA_io="romio321"
export UCX_HANDLE_ERRORS=bt
ulimit -s 102400
ulimit -c 0
srun -l --cpu_bind=cores --distribution=block:cyclic --propagate=STACK,CORE ./icon.exe
ENDFILE
This concludes the runfile. The Job_ICON-file that has been produced in the last step can then be executed. For that, the runfile hast to be run using bash:
$ chmod +x myrunfile
$ ./myrunfile
This will then create all the Namelist files and the runscript job_ICON. To submit the icon run, go to the output directory, make the runscript executable, and run it:
$ cd /path/to/your/output
$ chmod +x job_ICON
$ sbatch job_ICON
Now the Job should be submitted. The submission status can be seen with
$ squeue --user your_username
If everything went well, in the output directory there will be output files generated with names like Raikoke-June-2019-forecast_mode-remap_DOM01_ML_0001.nc
Inspecting the Output
In this example, the output is created in the form of several NetCDF4 (.nc) files. To avoid having one giant file, it is subdivided into several files containing a fixed amount of timesteps. The quickest way to inspect a NetCDF file is with CDO:
$ cdo sinfo Raikoke-June-2019-forecast_mode-remap_DOM01_ML_0001.nc
This way it is possible to obtain information about the variables, grid parameters and timesteps of the simulation. To actually see the values of a specific variable, tools like ncdump can be used, however in many cases this is not practical, since the amount of single values is way too large to look at individual data points.
Making a Plot of the Simulation Data
In order to gain insight into the simulation, as well as an overview, the most common way is to create some plots of different variables of interest. In our case, we want to know what happens to the ash emitted from the Raikoke eruption. A quick and easy way to create plots is using python and a reference script to produce a plot similar to the Pointsource animation is given here:
# first we import the following packages:
import numpy as np # general utility for trigonomic functions and arrays
import matplotlib.pyplot as plt # Library to produce Plots
import xarray as xr # reading in of netCDF data, optimized for large datasets
from glob import glob
import cartopy.crs as ccrs # plotting things on a map
#Next step: read in the Netcdf Files
#datadir_externally = 'path/to/your/output/Training2023/Volcano_externally/'
files_externally = glob(datadir_externally+'Raikoke-June-2019-forecast_mode-remap_DOM01_ML*')
files_externally.sort()
#Next step: Calculate relevant data to be displayed
# get model level height
def get_dz(ds):
dz = -1 * ds.z_ifc.diff('height_2')
dz = dz.rename({'height_2':'height'})
dz = dz.assign_coords(height=(dz.height - 1))
return dz
# Calculate column integrated tracer mass (tracer load)
def tracer_load(tr,rho,dz):
m_tr = tr * rho * dz
m_tr = m_tr.sum('height')
m_tr = m_tr.squeeze()
return m_tr
#
MM_SO2 = 64/28.96 # molar weight of SO2 / dry air
# set the timestep you want to plot
# in this simulation there are 144 timesteps and 144 files each containing one timestep
# To produce a plot for each timestep, you can also loop the rest of this file over i
i = 30
#load the NetCDF file containing the timestep
ds = xr.open_dataset(files_externally[i],autoclose=True)
#calculate Heightlevels
dz = get_dz(ds)
# Read in SO2 Column
SO2 = tracer_load(ds.TRSO2_chemtr.values*MM_SO2,ds.rho,dz)
# set colorbar levels
color_so2 = np.linspace(1e-5, 0.02, 11,endpoint=True)
# Plot SO2 column
# set the size of the figure and the background color to white
fig = plt.figure(figsize=(8,6),facecolor='w')
# Set the axes using the specified map projection
ax=plt.axes(projection=ccrs.NearsidePerspective(central_longitude=180, central_latitude=49.29,satellite_height=35785831/5, globe=None))
ax.stock_img() #add a stock image as background
ax.coastlines() # add coastlines
ax.set_title(str(ds.time.values)[2:12]+" "+str(ds.time.values)[13:18]) #set title to the current time displayed
ax.gridlines(draw_labels=False, dms=False, x_inline=True, y_inline=True) #add lat lon gridlines
# Make a filled contour plot
plot1=ax.contourf(ds.lon, ds.lat, SO2, levels=color_so2, cmap='plasma',
transform = ccrs.PlateCarree())
kwargs = {'format': '%.3f'}
cbar = plt.colorbar(plot1,location='right',ticks=color_so2,**kwargs)
cbar.set_label('SO2 Column Mass (kg m-2)')
#save the plot at a path of your choice
#plt.savefig("path/to/your/Plots/folder/SO2_{:03d}.png".format(i))
plt.close(fig)
To run the script, you need a working python version and the packages listed in the beginning of the script. Dont forget to make the file executable in order to run it using chmod +x plotscript .
The resulting Plot will look something like this: